Note
Go to the end to download the full example code
Laminar model comparison using cross-validation error#
This tutorial demonstrates how to perform laminar inference using model comparison based on cross-validation error as a metric of model fit, described in [Bonaiuto et al., 2018, Non-invasive laminar inference with MEG: Comparison of methods and source inversion algorithms](https://doi.org/10.1016/j.neuroimage.2017.11.068). A 20Hz oscillation is simulated at a particular cortical location in various layers. Source reconstruction is performed using the Empirical Bayesian Beamformer on the simulated sensor data using forward models based on different layer meshes. These models are then compared using cross-validation error. Cross-validation error is computed by fitting the model N times, each time leaving out a certain percentage of the channels and seeing how well the resulting model can predict the signal in those channels
Setting up the simulations#
Simulations are based on an existing dataset, which is used to define the sampling rate, number of trials, duration of each trial, and the channel layout.
import os
import shutil
import numpy as np
import matplotlib.pyplot as plt
import tempfile
from lameg.invert import coregister, invert_ebb
from lameg.simulate import run_current_density_simulation
from lameg.laminar import model_comparison
from lameg.surf import LayerSurfaceSet
from lameg.util import get_fiducial_coords
from lameg.viz import show_surface
import spm_standalone
# Subject information for data to base the simulations on
subj_id = 'sub-104'
ses_id = 'ses-01'
# Fiducial coil coordinates
fid_coords = get_fiducial_coords(subj_id, '../test_data/participants.tsv')
# Data file to base simulations on
data_file = os.path.join(
'../test_data',
subj_id,
'meg',
ses_id,
f'spm/spm-converted_autoreject-{subj_id}-{ses_id}-001-btn_trial-epo.mat'
)
spm = spm_standalone.initialize()
For source reconstructions, we need an MRI and a surface mesh. The simulations will be based on a forward model using the multilayer mesh, and the model comparison will use each layer mesh
surf_set_bilam = LayerSurfaceSet(subj_id, 2)
surf_set = LayerSurfaceSet(subj_id, 11)
We’re going to copy the data file to a temporary directory and direct all output there.
# Extract base name and path of data file
data_path, data_file_name = os.path.split(data_file)
data_base = os.path.splitext(data_file_name)[0]
# Where to put simulated data
tmp_dir = tempfile.mkdtemp()
# Copy data files to tmp directory
shutil.copy(
os.path.join(data_path, f'{data_base}.mat'),
os.path.join(tmp_dir, f'{data_base}.mat')
)
shutil.copy(
os.path.join(data_path, f'{data_base}.dat'),
os.path.join(tmp_dir, f'{data_base}.dat')
)
# Construct base file name for simulations
base_fname = os.path.join(tmp_dir, f'{data_base}.mat')
Invert the subject’s data using the multilayer mesh. This step only has to be done once - this is just to compute the forward model that will be used in the simulations
# Patch size to use for inversion (in this case it matches the simulated patch size)
patch_size = 5
# Number of temporal modes to use for EBB inversion
n_temp_modes = 4
# Coregister data to multilayer mesh
coregister(
fid_coords,
base_fname,
surf_set,
spm_instance=spm
)
# Run inversion
[_,_] = invert_ebb(
base_fname,
surf_set,
patch_size=patch_size,
n_temp_modes=n_temp_modes,
spm_instance=spm
)
Simulating a signal on the pial surface#
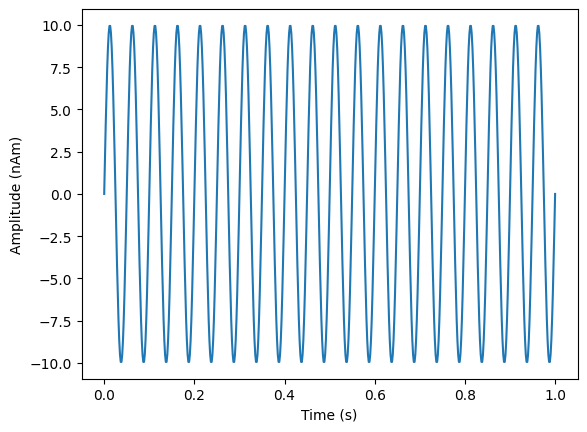
We’re going to simulate 1s of a 20Hz sine wave with a dipole moment of 10nAm
# Frequency of simulated sinusoid (Hz)
freq = 20
# Strength of simulated activity (nAm)
dipole_moment = 10
# Sampling rate (must match the data file)
s_rate = 600
# Generate 1s of a sine wave at a sampling rate of 600Hz (to match the data file)
time = np.linspace(0,1,s_rate+1)
sim_signal = np.sin(time*freq*2*np.pi).reshape(1,-1)
plt.plot(time,dipole_moment*sim_signal[0,:])
plt.xlabel('Time (s)')
plt.ylabel('Amplitude (nAm)')
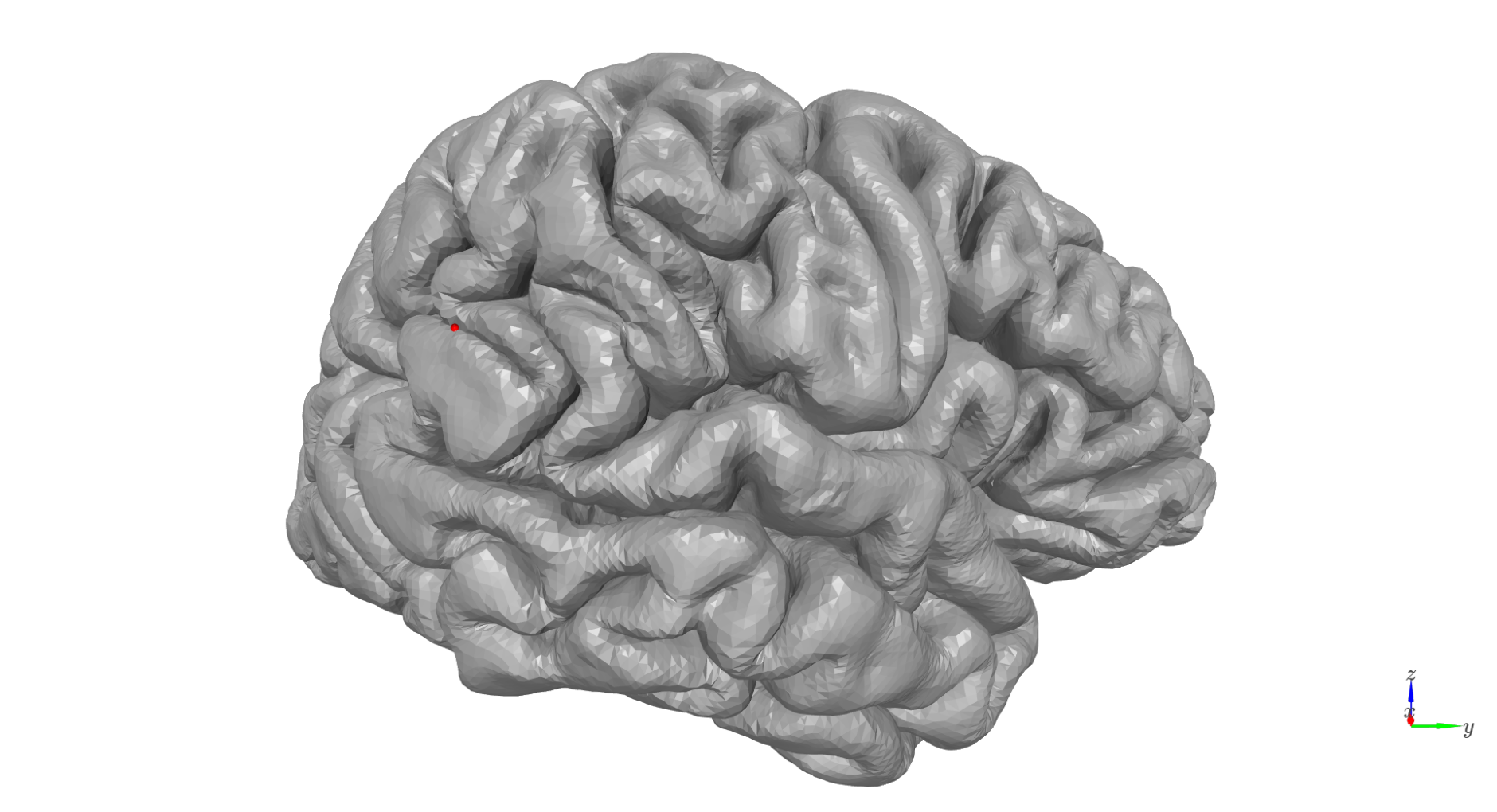
We need to pick a location (mesh vertex) to simulate at
# Vertex to simulate activity at
sim_vertex=10561
cam_view = [40, -240, 25,
60, 37, 17,
0, 0, 1]
plot = show_surface(
surf_set,
marker_vertices=sim_vertex,
marker_size=5,
camera_view=cam_view
)
We’ll simulate a 5mm patch of activity with -5 dB SNR at the sensor level. The desired level of SNR is achieved by adding white noise to the projected sensor signals
# Simulate at a vertex on the pial surface
pial_vertex = surf_set.get_multilayer_vertex('pial', sim_vertex)
prefix = f'sim_{sim_vertex}_pial_'
# Size of simulated patch of activity (mm)
sim_patch_size = 5
# SNR of simulated data (dB)
SNR = -5
# Generate simulated data
pial_sim_fname = run_current_density_simulation(
base_fname,
prefix,
pial_vertex,
sim_signal,
dipole_moment,
sim_patch_size,
SNR,
spm_instance=spm
)
Model comparison (pial - white matter)#
Now we can run model comparison between source models based on the pial and white matter surfaces using cross-validation error. For computing cross-validation error, we’ll run 10 folds, leaving out 10% of the channels in each fold. We’ll then look at the difference in cross-validation error between the two models (pial - white matter). This should be lower for the pial surface model because we simulated activity on the pial surface
# Number of cross validation folds
n_folds = 10
# Percentage of test channels in cross validation
ideal_pc_test = 10 # may not use this number as we need integer number of channels
# Run model comparison between the first layer (pial) and the last layer (white matter)
[_,cvErr] = model_comparison(
fid_coords,
pial_sim_fname,
surf_set_bilam,
spm_instance=spm,
invert_kwargs={
'patch_size': patch_size,
'n_temp_modes': n_temp_modes,
'n_folds': n_folds,
'ideal_pc_test': ideal_pc_test
}
)
The difference in cross validation error after averaging over test channels and folds This value should be negative (less error for the pial layer model)
np.mean(np.mean(cvErr[0],axis=-1),axis=-1)-np.mean(np.mean(cvErr[1],axis=-1),axis=-1)
White matter surface simulation with pial - white matter model comparison#
Let’s simulate the same pattern of activity, in the same location, but on the white matter surface. This time, model comparison should yield lower cross-validation error for the white matter surface.
# Simulate at the corresponding vertex on the white matter surface
white_vertex = surf_set.get_multilayer_vertex('white', sim_vertex)
prefix = f'sim_{sim_vertex}_white_'
# Generate simulated data
white_sim_fname = run_current_density_simulation(
base_fname,
prefix,
white_vertex,
sim_signal,
dipole_moment,
sim_patch_size,
SNR,
spm_instance=spm
)
# Run model comparison between the first layer (pial) and the last layer (white matter)
[_,cvErr] = model_comparison(
fid_coords,
white_sim_fname,
surf_set_bilam,
spm_instance=spm,
invert_kwargs={
'patch_size': patch_size,
'n_temp_modes': n_temp_modes,
'n_folds': n_folds,
'ideal_pc_test': ideal_pc_test
}
)
The difference in cross validation error after averaging over test channels and folds This value should be positive (less error for the white matter layer model)
np.mean(np.mean(cvErr[0],axis=-1),axis=-1)-np.mean(np.mean(cvErr[1],axis=-1),axis=-1)
Simulation in each layer with model comparison across layers#
That was model comparison with two candidate models: one based on the white matter surface, and one on the pial. Let’s now simulate on each layer, and for each simulation, run model comparison across all layers. We’ll turn off SPM visualization here.
# Now simulate at the corresponding vertex on each layer, and for each simulation, run model comparison across
# all layers
all_layerCvErr = []
for l in range(surf_set.n_layers):
print(f'Simulating in layer {l}')
l_vertex = surf_set.get_multilayer_vertex(l, sim_vertex)
prefix = f'sim_{sim_vertex}_{l}_'
l_sim_fname = run_current_density_simulation(
base_fname,
prefix,
l_vertex,
sim_signal,
dipole_moment,
sim_patch_size,
SNR,
spm_instance=spm
)
[_,layerCvErr] = model_comparison(
fid_coords,
l_sim_fname,
surf_set,
viz=False,
spm_instance=spm,
invert_kwargs={
'patch_size': patch_size,
'n_temp_modes': n_temp_modes,
'n_folds': n_folds,
'ideal_pc_test': ideal_pc_test
}
)
all_layerCvErr.append(layerCvErr)
all_layerCvErr = np.array(all_layerCvErr)
# Average over test channels and folds
all_layerCvErr = np.mean(np.mean(all_layerCvErr, axis=-1), axis=-1)
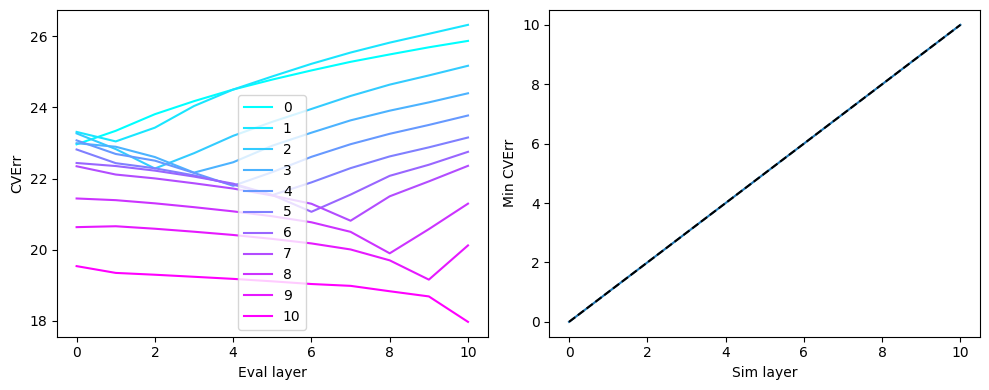
For each simulation, we can plot the cross-validation error for all models relative to the worst model. The layer model with the lowest cross-validation error should correspond to the layer that the activity was simulated in.
col_r = plt.cm.cool(np.linspace(0,1, num=surf_set.n_layers))
plt.figure(figsize=(10,4))
# For each simulation, plot the CV error of each layer model relative to that of the worst
# model for that simulation
plt.subplot(1,2,1)
for l in range(surf_set.n_layers):
layerCvErr=all_layerCvErr[l,:]
plt.plot(layerCvErr, label=f'{l}', color=col_r[l,:])
plt.legend()
plt.xlabel('Eval layer')
plt.ylabel('CVErr')
# For each simulation, find which layer model had the lowest CV error
plt.subplot(1,2,2)
peaks=[]
for l in range(surf_set.n_layers):
layerCvErr=all_layerCvErr[l,:]
pk=np.argmin(layerCvErr)
peaks.append(pk)
plt.plot(peaks)
plt.xlim([-0.5,10.5])
plt.ylim([-0.5,10.5])
plt.plot([0,10],[0,10],'k--')
plt.xlabel('Sim layer')
plt.ylabel('Min CVErr')
plt.tight_layout()
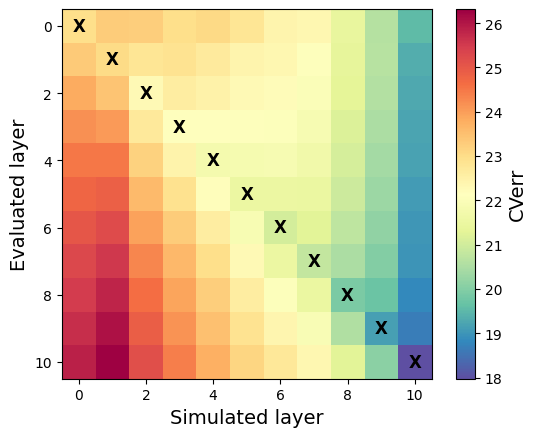
# Transpose for visualization
im=plt.imshow(all_layerCvErr.T, cmap='Spectral_r')
# Find the indices of the min value in each column
min_indices = np.argmin(all_layerCvErr, axis=1)
# Plot an 'X' at the center of the square for each column's minimum
for idx, min_idx in enumerate(min_indices):
plt.text(idx, min_idx, 'X', fontsize=12, ha='center', va='center', color='black', weight='bold')
plt.xlabel('Simulated layer', fontsize=14)
plt.ylabel('Evaluated layer', fontsize=14)
cb=plt.colorbar(im)
cb.set_label('CVerr', fontsize=14)
spm.terminate()
# Delete simulation files
shutil.rmtree(tmp_dir)
Total running time of the script: (0 minutes 0.000 seconds)